Research
Dynamic characterization and interpretation for protein–RNA interactions across diverse cellular conditions using HDRNet
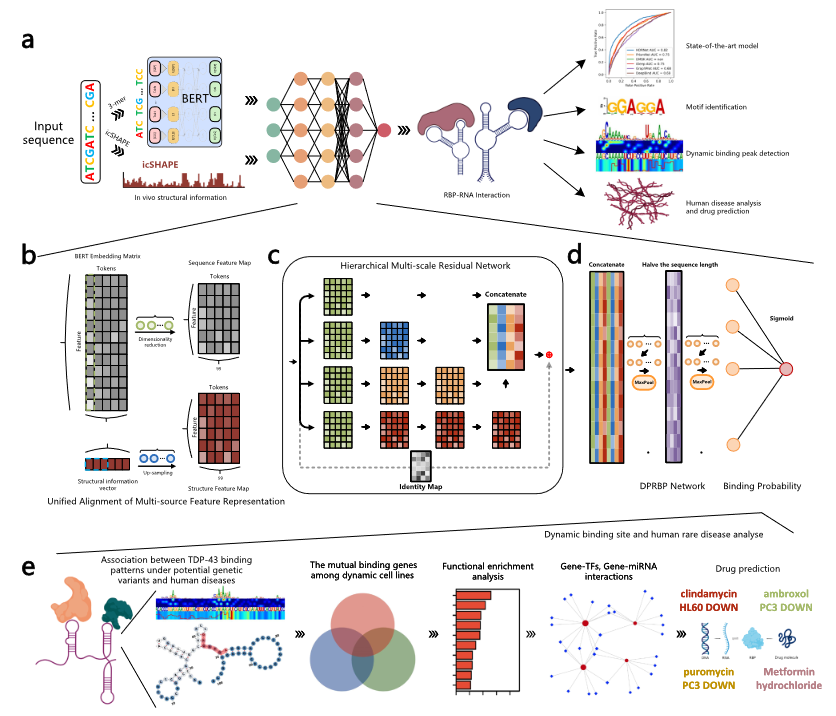
Here, we develop HDRNet, an end-to-end deep learning-based framework to precisely predict dynamic RBP binding events under diverse cellular conditions. Our results demonstrate that HDRNet can accurately and efficiently identify binding sites, particularly for dynamic prediction, outperforming other state-of-the-art models on 261 linear RNA datasets from both eCLIP and CLIP-seq, supplemented with additional tissue data. Moreover, we conduct motif and interpretation analyses to provide fresh insights into the pathological mechanisms underlying RNA-RBP interactions from various perspectives. Our functional genomic analysis further explores the gene-human disease associations, uncovering previously uncharacterized observations for a broad range of genetic disorders.
H. Zhu, Y. Yang, Y. Wang, F. Wang, Y. Huang, Y. Chang, K. Wong*, X. Li*
Nature Communications(2023)
Topological Identification and Interpretation for Single-cell Gene Regulation Elucidation across Multiple Platforms using scMGCA

We develop a deep graph learning method, scMGCA, for single-cell data analysis. scMGCA is based on a graph-embedding autoencoder that simultaneously learns cell-cell topology representation and cluster assignments. We show that scMGCA is accurate and effective for cell segregation and batch effect correction, outperforming other state-of-the-art models across multiple platforms. In addition, we perform genomic interpretation on the key compressed transcriptomic space of the graph-embedding autoencoder to demonstrate the underlying gene regulation mechanism. We demonstrate that in a pancreatic ductal adenocarcinoma dataset, scMGCA successfully provides annotations on the specific cell types and reveals differential gene expression levels across multiple tumor-associated and cell signalling pathways.
Z. Yu, Y. Su, Y. Lu, F. Wang, S. Zhang, Y. Chang, K. Wong*, X. Li*
Nature Communications(2023)
Distribution-Agnostic Deep Learning Enables Accurate Single-Cell Data Recovery and Transcriptional Regulation Interpretation
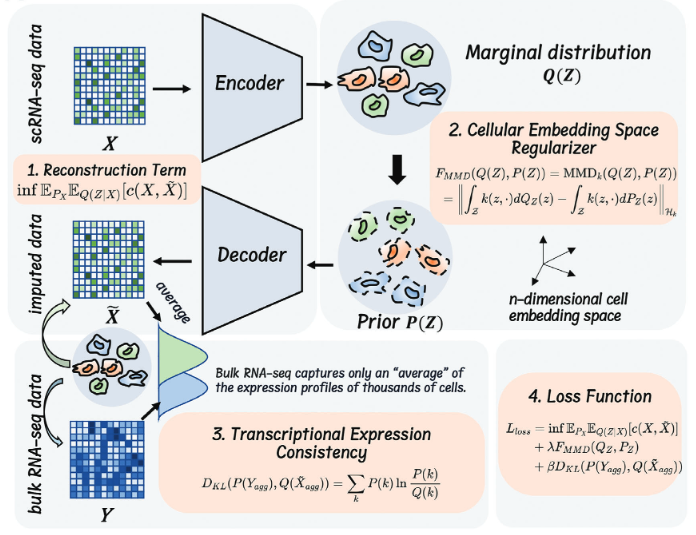
We developed Bis, a distribution-agnostic deep learning model for accurately recovering missing sing-cell gene expression from multiple platforms. Bis is an optimal transport-based autoencoder model that can capture the intricate distribution of scRNA-seq data while addressing the characteristic sparsity by regularizing the cellular embedding space. Additionally, they propose a module using bulk RNA-seq data to guide reconstruction and ensure expression consistency. Experimental results show Bis outperforms other models across simulated and real datasets, showcasing superiority in various downstream analyses including batch effect removal, clustering, differential expression analysis, and trajectory inference. Moreover, Bis successfully restores gene expression levels in rare cell subsets in a tumor-matched peripheral blood dataset, revealing developmental characteristics of cytokine-induced natural killer cells within a head and neck squamous cell carcinoma microenvironment.
Y. Su, Z. Yu, Y. Yang, Y. Huang, K. Wong, X. Li*
Advanced Science(2024)
Reliable Identification and Interpretation of Single-cell Molecular Heterogeneity and Transcriptional Regulation using Dynamic Ensemble Pruning

A dynamic ensemble pruning framework (DEPF) is proposed to identify and interpret single-cell molecular heterogeneity. In particular, a silhouette coefficient-based indicator is developed and evaluated to determine the optimization direction of the bi-objective function. Following that, a bi-objective fruit fly optimization algorithm is designed to prune dynamically the low-quality basic clustering in the ensemble.
Y. Fan, Y. Wang, F. Wang, L. Huang, Y. Yang, K. Wong, X. Li*
Advanced Science(2023)
Enabling Single-cell Drug Response Annotations from Bulk RNA-seq using SCAD

A transfer learning model is proposed to infer the drug sensitivities at single-cell level. This framework learns bulk transcriptome profiles and pharmacogenomics information from population cell lines in a large public dataset and transfers the knowledge to infer drug efficacy of individual cells. The results suggest that it is suitable to learn knowledge from pre-clinical cell lines to infer pre-existing cell subpopulations with different drug sensitivities prior to drug exposure. In addition, the model offers a new perspective on drug combinations. It is observed that drug-resistant subpopulation can be sensitive to other drugs (e.g., a subset of JHU006 is Vorinostat-resistant while Gefitinib-sensitive); such finding corroborates the previously reported drug combination (Gefitinib + Vorinostat) strategy in several cancer types. The identified drug sensitivity biomarkers reveal insights into the tumor heterogeneity and treatment at cellular resolution.
Z. Zheng, J. Chen, X. Chen, L. Huang, W. Xie, Q. Lin, X. Li*, K. Wong*
Advanced Science(2023)
A lightweight framework for chromatin loop detection on single-cell Hi-C

A lightweight framework called scGSLoop is proposed, which sets a new paradigm for scHi-C loop calling by adapting the training and inferencing strategies of graph-based deep learning to leverage the sequence features and 1D positional information of genomic loci. With this framework, sparsity is no longer a challenge, but rather an advantage that the model leverages to achieve unprecedented computational efficiency. Compared to existing methods, scGSLoop makes more accurate predictions and is able to identify more loops that have the potential to play regulatory roles in genome functioning. Moreover, scGSLoop preserves single-cell information by identifying a distinct group of loops for each individual cell, which not only enables an understanding of the variability of chromatin looping states between cells, but also allows scGSLoop to be extended for the investigation of multi-connected hubs and their underlying mechanisms.
F. Wang, H. Alinejad-Rokny, J. Lin, T. Gao, X. Chen, L. Meng, X. Li*, K. Wong*